What is PCR?
Introduction
Polymerase Chain Reaction (PCR) is a laboratory method that facilitates the quick duplication of particular DNA sequences. By constantly undergoing temperature fluctuations, the PCR process generates millions of replicas of a specific gene or DNA fragment. This technology holds considerable value in research, diagnostics, and forensic science.
Principle of PCR
The core concept of PCR is that when a double-stranded DNA molecule is subjected to high temperatures, it unwinds into two single-stranded DNA molecules. These single strands can be readily replicated using DNA polymerase and nucleosides, leading to the duplication of the initial DNA strands. By continuously repeating this process, numerous copies of the original DNA molecule can be produced.
History of PCR

The polymerase chain reaction (PCR) was created in 1983 by Dr. Kary Mullis during his tenure at Cetus Corporation. He was awarded the Nobel Prize in Chemistry in 1993 for this groundbreaking innovation that transformed molecular biology. This technique enables the amplification of DNA sequences from various organisms. Over the years, it has been modified to facilitate the amplification of RNA samples, as well as to quantify the levels of DNA or RNA present in a sample. The discovery of a heat-stable DNA polymerase (Taq) from an archaebacterium sourced from a geothermal vent in Yellowstone National Park made it possible to perform the reaction in a single closed tube using different temperatures.
Purpose of PCR
PCR is an exceptionally effective technique for the specific in vitro amplification of nucleic acids. Its numerous applications highlight its usefulness. The power of PCR lies in the minimal amount of initial material needed. Adjusting specificity can be accomplished by simply changing the length and nucleotide sequence of primers and the annealing temperature. This flexibility is especially critical in medical diagnostics when an infectious agent is present in small quantities. Additionally, PCR is a key diagnostic tool for various genetic disorders and chimerism assessments in bone marrow transplants. Moreover, it has been crucial in studying microbial species, such as amplifying and sequencing 16S rRNA to explore the evolutionary relationships among diverse bacterial species.
Requirements of PCR Technique
Material for PCR Technique
- DNA template:
- DNA template contains the specific DNA region (target) that is to be amplified, serving as the source of DNA for the PCR process. In the laboratory, a standard concentration of 25ng/μl is typically utilized. The purity of the DNA is crucial, as contaminants (such as excess phenolic compounds or EDTA) can inhibit PCR and lead to false-negative outcomes.
- Primer pairs:
- Short synthetic DNA fragments harboring sequences that are complementary to the target region; these correspond to the 3′ (three prime) ends of the sense (forward) strand and the 5′ end of the anti-sense (reverse) strand of the DNA target, usually consisting of 18-30 nucleotides in length.
- Master Mix includes:
- Taq DNA polymerase, the enzyme responsible for joining the free nucleotides. It begins at the 3′ end of the primer and utilizes the complementary DNA strand as a template.
- Deoxynucleoside triphosphates (dNTPs, also referred to as “deoxynucleotide triphosphates”); these are the free nucleotides (G, A, T, C) that serve as the building blocks for the synthesis of new DNA strands by the polymerase.
- A buffer solution to help maintain the pH and ionic strength of the reaction, making conditions suitable for the enzyme’s activity.
- Mg++ ions, which act as a cofactor for the enzyme.
- Nuclease-free water
- Which is essential for the reaction to proceed effectively.
Protocol of PCR Technique
Usually 20 to 50 μl total in volume and include the following:
- 0.1 to 1 μg of genomic DNA or cDNA, ~0.1μg should be sufficient for plasmid DNA.
- 10X PCR buffer to give a final concentration of 1X
- 4 mM dNTP mix (dCTP, dATP, dGTP, dTTP) to give a final concentration of 0.2 mM
- Both the forward and reverse primer added at a final concentration of 0.1 μM to 1 μM of each primer
- 1 unit/μl Taq polymerase
- H2O (DNA and DNase free) to bring volume to 20 μl to 50 μl
An example 20 μl reaction
- 1 μl of dsDNA template (~0.1 μg)
- 2 μl of 10X buffer
- 1 μl of 4 mM dNTP mix
- 1 μl of 10 μM forward primer to a final concentration of 0.5 μM
- 1 μl of 10 μM reverse primer to a final concentration of 0.5 μM
- 1 μl of 1 unit/μl Taq polymerase
- 13 μl of water
Combine the reagents in either a 0.5-ml tube or a 0.2-ml PCR tube. Ensure that the reagents remain on ice. Gently tap the tube to mix the contents and briefly spin it in a microcentrifuge to collect everything at the bottom, then keep it on ice until you are ready to load it into the thermocycler. If the thermocycler lacks a heated lid, add a thin layer of mineral oil on top of the mixture to reduce evaporation during cycling.
How to reduce the chance for contamination during PCR
PCR enables the production of over 10 million copies of a target DNA sequence starting from just a few molecules. Consequently, PCR is highly susceptible to contamination from non-target DNA. To minimize the risk of contamination, several precautions should be taken, including:
- Fresh gloves must be worn for DNA purification and during the setup of each reaction.
- Employing aerosol tips (those with a cotton wad at the top) is advisable.
- Minimize direct contact with sample tubes and reagents; for example, avoid spitting in your workspace.
- The processes of DNA sample preparation, assembling the reaction mixture, and carrying out the PCR, as well as the analysis of the resulting products, should be conducted in distinct areas.
- The reagents intended for PCR should be exclusively prepared for this application.
- Only nuclease-free water must be utilized in the preparation and dilution of PCR reagents.
- Autoclaving all solutions, except for dNTPs, primers, and Taq DNA polymerase, is recommended unless the solution is already acquired as sterile.
- Solutions should be aliquoted into small portions and stored in designated PCR areas, with aliquoted PCR reagents kept separate from DNA samples.
The PCR reaction
Thermalcycler
A thermal cycler, often referred to as a PCR machine or DNA amplifier, is a laboratory device utilized to amplify DNA segments through the polymerase chain reaction (PCR). The thermal cycler heats and cools the reaction tubes. The thermal cycler (also known as a thermocycler, PCR machine or DNA amplifier) is a laboratory apparatus most commonly used to amplify segments of DNA via PCR).Thermal cyclers may also be used in laboratories to facilitate other temperature-sensitive reactions, including restriction enzyme digestion or rapid diagnostics
Key Components and Structure of Thermalcycler
Typically, a thermal cycler is comprised of the following main components:
- Heating Block:
- The heating block serves as the primary component where the PCR tubes or plates are positioned. It is engineered to rapidly and evenly adjust temperatures to support different phases of the PCR process. The block includes wells or slots for holding the reaction vessels.
- Peltier Elements:
- These thermoelectric components allow for precise temperature regulation by heating or cooling the block. They enable the thermal cycler to transition through the necessary temperatures for denaturation, annealing, and extension.
- Temperature Sensors:
- These sensors continuously monitor the block’s temperature in real-time, ensuring precise and consistent conditions throughout the PCR procedure.
- Control Interface:
- Contemporary thermal cyclers are equipped with user-friendly controls, often featuring touchscreens or software applications, for programming and overseeing the PCR cycles. Users can customize parameters such as temperature, duration, and the total number of cycles.
- Lid Heater:
- The lid of the thermal cycler incorporates a heating element to avoid condensation forming on the inner surface of the tube caps, which could otherwise change the reaction volume and impact the results.
- Cooling System:
- Some thermal cyclers come with a cooling mechanism to quickly reduce the block’s temperature when necessary, enhancing the efficiency of the cycling process.
Example typical thermal cycler program
- Step 1: 92 to 98 C, 30 seconds to 1 minute
- Step 2: optimal annealing temperature of primers, 37 to 65 C, 30 seconds to 1 minute
- Step 3: 72 C, 30 seconds to 1 minute
- Repeat steps 1 to 3 for 20 to 30 times to accumulate enough amplified target DNA to be visualized on a gel.
- Step 4: 4 C holding of sample until analysis by gel electrophoresis
Procedure (Steps) of PCR Technique
Initialization step:
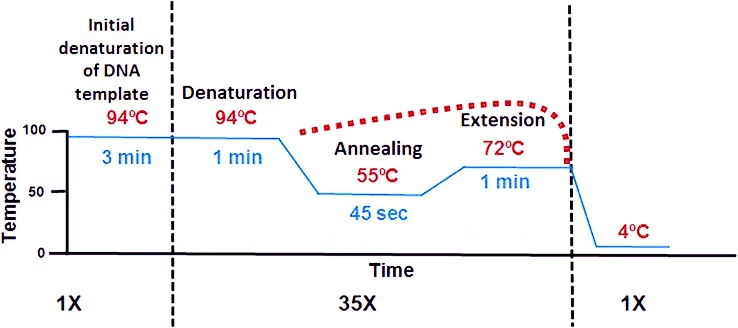
This step consists of heating the reaction to a temperature of 94–96 °C (or 98 °C if extremely thermostable polymerases are used), which is held for 1–9 minutes. It is only required for DNA polymerases that require heat activation by hot-start PCR. PCR involves a series of temperature cycles that consist of three primary stages:
Denaturation (94-98°C):
This step is the first regular cycling event and consists of heating the reaction chamber to 94–98 °C for 20–60 seconds. This causes DNA melting or denaturation, of the double-stranded DNA template by breaking the hydrogen bonds between complementary bases, yielding two single-stranded DNA molecules.

Annealing (50-65°C):
- The reaction temperature is lowered to 50–65 °C (122–149 °F) for 20–40 seconds
- Ionic bonds are constantly formed and broken between the single stranded primer and the single stranded template.
- The more stable bonds last a longer (primers that fit exactly) ,the polymerase can attach and starts copying the template.
Extension or Elongation (72°C):
- The optimal temperature for Taq polymerase is 72°C. The polymerase extends the primers in the 5′ to 3′ direction.
- The extension time depends upon the length of the target DNA sequence but a general rule of thumb is one thousand bases per minute at optimal temperature.
- The temperature that is used during the extension phase is dependent on the DNA polymerase that is used
Final elongation:
This single step is occasionally performed at a temperature of 70–74 °C for 5–15 minutes after the last PCR cycle to ensure that any remaining single-stranded DNA is fully extended.
These steps are repeated for 20-40 cycles, leading to exponential amplification of the target DNA sequence.
Detection of PCR Products
PCR products are identified using agarose gel electrophoresis, which sorts DNA fragments by their size. Additional techniques involve employing probes and dyes to visualize the PCR products.
Agarose gel electrophoresis
- An electric current drives DNA fragments through a gel medium.
- Shorter DNA strands travel more quickly than longer ones.
- The arrangement of DNA fragments is evaluated against known controls.
- The brightness of the bands can provide an estimate of the product quantity.
- Ethidium bromide is a widely used stain for visualizing DNA products.
- SYBR green is a safe dye that is also applicable for visualizing DNA products.
Probes and dyes
- Scorpion primers are specific probes that merge a PCR primer with a DNA probing sequence.
- Molecular beacons function similarly to Scorpion primers.
- 6-Carboxyfluorescein (FAM) and tetrachloro-6-carboxyfluorescein (TET) are fluorescent dyes utilized to detect specific alleles.
Applications of PCR
- Genome study
- Quantitation of gene expression
- DNA damage (microsatellite instability) measurement
- Detection of inactivation of gene at X- chromosome
- Genotyping
- Microbes detection
- Viral quantitation
- Bacterial
- Monitoring
- Drug therapy efficacy / drug monitoring
- Prenatal diagnosis
- Cancer diagnostics
- Forensic Science:
- DNA fingerprinting for criminal investigations and paternity testing.
- Archaeology and Anthropology:
- Studying ancient DNA and human evolution.
- Environmental Science:
- Detecting and quantifying microbial communities in environmental samples.
- Agriculture:
- Genetically modified organism (GMO) detection and plant breeding.
